Libraries tutorial
1. Logging in
You need to log in to MISO LIMS in order to make changes to any LIMS entities. Logging in lets MISO record any changes you make on entities you have access to.
If you are a new user, you will need
to contact helpdesk@oicr.on.ca so that they put you into the appropriate
Active Directory group, MISO_ROLE_INTERNAL.
Try to log in now:
- Click on http://miso.gsi.oicr.on.ca.
- Enter your username (e.g. jdoe) and password and click the Login button.
MISO uses the same username and password as your OICR email account.
If all goes well, you should see the MISO Dashboard and see a message at the top right: "Logged in as: jdoe".
2. Propagating aliquots to libraries
A library is made from one sample for a single target platform and has a specific design associated with it that decides the selection and strategies used to make the library. A library may also have indices (primers/sequencing barcodes/molecular IDs) and QC information.
MISO stores two important pieces of information about how a library was generated: the selection (e.g., PCR, cDNA) and the strategy (e.g., WGS, WXS, amplicon). A design captures both a selection and strategy and the list of allowed designs is limited based on the sample type (e.g., a cDNA sample can only have SM, WT, or MR library designs and these lock the selection and strategy type accordingly).
2.0 Scan aliquots into your inbox
First, we accept the aliquots made by the samples team and put them into your inbox.
Boxes hold samples, libraries, library aliquots, and pools. They do not track reagents or primers. Boxes are separated into uses for different kinds of storage (e.g., tissue samples versus extracted DNA), but there is no check that items in a box match. Each box also has a size that includes the physical dimensions of the box as well as whether the box is compatible with the VisionMate scanner. Every position in the box is identified by a standard row letter + column number format (e.g., C05).
In this exercise, we are going to take samples/libraries/pools from the previous team/collaborator by scanning them out of the previous box and into a newly created 'inbox'.
If you haven't already created an inbox, create one now:
- From the navigation panel, choose Boxes.
- Click Add at the top right of the table.
- In the Create Boxes dialog, leave the Quantity at 1 and click Create.
- On the Create Box page, enter the following information:
- Alias:
XX Inbox, replacingXXwith your own initials. Record this alias in your worksheet.
- Description: a human description of the box purpose
gDNA ready for library prep for <project>. - Box Use: The contents of the box. Select an appropriate use for your entities.
Record the use in your worksheet.

- Box Size: Select
8x12 Storage (scannable).
- Alias:
- Click Save.
Upon clicking save, a graphic of the box will appear.
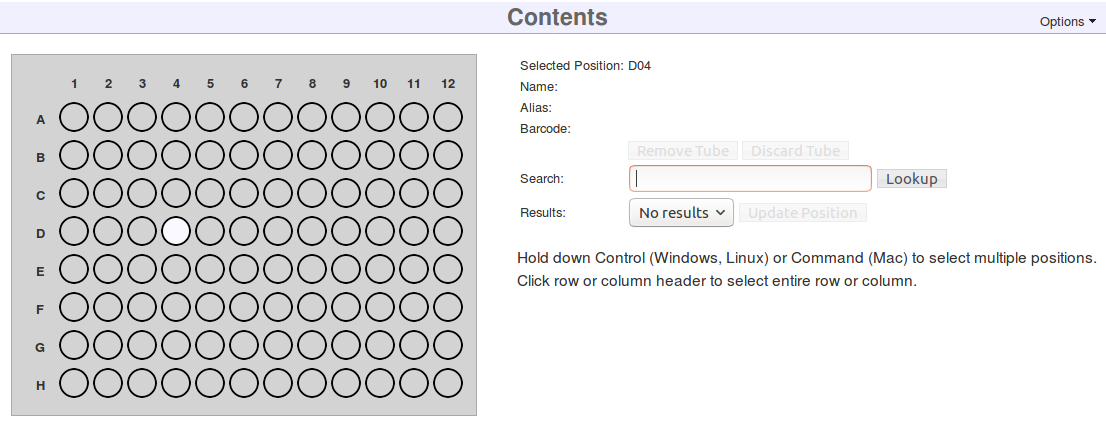
You can now fill the box with the samples, libraries, library aliquots, or pools that were made in previous steps. Collect the appropriate matrix tubes/barcodes now.
- Click on a position in the displayed Contents grid.
- Enter a matrix barcode into the Search box on the right and click Lookup. Normally a hand scanner would be used. You can also search for items by their name or alias
- Click Update Position.
- ⚠ The Save button at the top of the page does not work for individual positions, only for Box Information.
- Repeat for as many samples/libraries/library aliquots/pools as you want.
The table below the box diagram shows the position and information for the currently selected sample. If you would like to see all of the samples in the table, click the List all Box Contents button at the top right of the table.
In the lab, it is possible to use Options > Scan Box to use the plate scanner and update all positions at once, but that will not be covered in this tutorial.
Using Boxes
Boxes can be found either from the Sample or Library page or the Boxes page.
- On the Samples or Libraries page, search for one of your sample or library aliases and click on its alias to load the page.
- The Box and position is listed under Box Location in the Sample Information
or Library Information section. Click on the link to go to the Box (e.g.
PROJ_OUTBOX, A03).
Boxes can be used to store Samples, Libraries, Library Aliquots, and Pools and one box can store all four types.
2.1 Bulk propagate aliquots into libraries
In this section, you will use the sample aliquots created already to create libraries.
-
On the Samples page, enter your project's short name into the search box and press the
Enterkey on your keyboard. -
Check the 4 gDNA (aliquot) samples created previously.
- Click the Propagate button at the top left of the table.
- In the dialog that appears, choose
1replicate. If necessary, choose to propagate toLibrary. - Click Propagate.
- A table will appear. Enter the library information:
- Library Name: Leave blank as this will be filled in automatically after save.
- Library Alias: Leave blank as this will be filled in automatically.
- Matrix Barcode: As before, usually this would be scanned by the hand scanner. In this tutorial, enter matrix
barcodes in the form
(project short name)_(row number)_lib, e.g.PROJ_1_lib. - SOP: Choose any value if required.
-
Thermal Cycler: Choose any value if required.
-
Design: Select any
- Platform:
Illumina -
Type:
Paired End - Index Kit:
Nextera DNA Dual Index - Index 1 and Index 2 (if available): Select any combination of indices you wish. Select different indices for each library. Selecting the same indices for two different libraries would make it unwise to pool those two libraries together later.
- Kit: Select any
- Kit Lot: Enter today's date, e.g.
2021-06-15 - QC Status:
Not Ready -
Size (bp):
300 - Volume:
100 - Vol. Units:
µL
- Choose Save. Record the Library aliases and barcodes in your worksheet.

Note that for dual-index libraries, only the first index needs to be specified. The second is optional.
2.2 Bulk propagate aliquots to libraries from Box page
In this section, you will use the aliquots created already and create libraries. These libraries can be sorted based on the parent sample's location within the box, and indices can be added based on the box position.
- On the Boxes page, find your box and click on its link.
- Select all the aliquots in your box by control-clicking all the aliquots in the box (command-click on a Mac). If the aliquots are all in one row or column, you can click on the row/column header to select all items in that row or column. Use control-click (or command-click) to select more than one row or column.
- Click the Propagate button at the top left of the box contents table. If the top left of the box contents table indicates that more than one type of element is selected, de-select the non-aliquot tubes and then click Propagate.
- In the dialog that appears, choose
1replicate. If necessary, choose to propagate toLibrary. - Click Propagate.
- Click the Sort button at the top of the table.
- In the dialog that appears, choose
Sample Location (by rows)as the Primary Sort. Leave all other options at their default values. - Click Sort. The libraries you are creating are now sorted based on their parent sample's position within the box.
Sorting can be done by rows (
A01,A02,B01,B02, …) or columns (A01,B01,A02,B02, …).- This may be useful when using the Sciclone robot to create libraries and add indices at a particular plate location. Once the libraries are sorted based on the parent sample's location, the indices can be copy-pasted in the same order that the robot requires.
- Do not save these libraries.
3. Receiving Libraries
Propagating libraries from samples is only possible if there are existing samples to propagate from, and only makes sense if the libraries were prepared in-house. Libraries may also be received without the samples. The sample information is still required, but it can all be entered in one step.
When a library is entered in this way, the automatically-created sample hierarchy will include an Identity, Tissue, Stock, and Aliquot. Existing Identities and Tissues may be included in the hierarchy if they match the data entered.
3.1 Entering received Libraries
- On the left hand menu under Preparation, click Libraries.
-
Click the Receive button at the top of the table.
-
In the dialog, select Aliquot Class:
gDNA (aliquot)and Quantity:1, and click Receive -
Select or enter the following data:
-
Project: Select your project from the drop-down.
-
Subproject: Select the subproject you created.
- Sample Type:
GENOMIC - Sci. Name:
Homo sapiens - External Name: Enter
PROJ_ID10, replacing thePROJwith your own project short name. The Identity Alias column should change to showFirst Receipt (PROJ). - Donor Sex: Select any.
- Tissue Origin:
Pa (Pancreas) - Tissue Type:
P (Primary Tumour) - Times Received:
1 - Tube Number:
1 - Timepoint:
Week 8 - Date of receipt: Select today's date
- Received From: Select any lab.
- Received By: Select any group.
-
Matrix Barcode: Enter
PROJ_lib5, replacingPROJwith your own project short name. -
Design: Select any.
- Platform:
Illumina -
Type:
Paired End - Index Kit:
No indices - Kit: Select any.
- QC Status:
Ready - Size (bp):
430
-
- Click Save at the top right. Record the Library alias and barcode in your worksheet.

3.2 Receipt Transfers
When receiving libraries, you can enter some receipt information including a date and time, the lab that the libraries came from, the internal group who was responsible for receiving them, a verification that they were received as expected, and that a QC check was performed on them. Entering this information causes a Transfer to be recorded. We will have a look at the transfer that was created from the data you entered in exercise 3.1.
- On the main menu under Preparation, click Libraries.
- Click the alias of a library you created in exercise 3.1. This takes you to the Edit Library page, which gives you a detailed view of the individual library and other items related to it.
- On the Edit Library page, scroll down to the Transfers section. You should see a single transfer in the list.
- Click the ID of the transfer to go to the Edit Transfer page, which gives you a more detailed view of the transfer.
- Scroll down and look at the Items section on the Edit Transfer page. If you create multiple items in the same project with the same date and time of receipt, they will be included in the same transfer.
Receipts are one type of transfer. You can also create internal transfers when items are transferred between different groups within your organization, and distribution transfers when items are transferred out of the organization. This allows you to track the complete chain of custody.
4. Quality control
There are three ways to indicate library quality in MISO: 1) Enter quantitative QC values under the Library QC section; 2) The overall pre-sequencing quality field QC Status; and 3) The post-sequencing quality control flag Low quality library.
4.1 Library QC
After measuring the insert size or concentration, this information can be entered into each library.
Several QC methods are supported in MISO. The same process can be used to add QCs to a single or multiple libraries at once.
- On the Libraries list page, enter your project's
name in the search box and press the
Enterkey on your keyboard. - Check the boxes for the libraries that you created in exercise 2.1.
- Click Add QCs at the top left of the table.
- Enter
1QC per library and1control per QC, and click Add. - Fill out the table for each row as follows:
- Date: enter today's date
- Type: Select one of the available QC methods.
- Instrument: Depending on the QC type, you may have to indicate which instrument was used. If required, chose any instrument.
- Kit: Depending on the QC type, you may have to indicate which kit was used. If required, choose any kit.
- Kit Lot: If you entered a kit, also include a LOT#. Use the current date. e.g.
2020-07-17 - Result: Enter a value that makes sense for the QC. Some QCs are pass/fail, while others have a numerical result.
- Control 1: Depending on the QC type, you may have to indicate which control(s) were used. If required, choose any control.
- Control 1 LOT#: If you entered a control, also include a LOT# for it. Use the current date. e.g.
2020-07-17. - Control 1 Passed?: If you entered a control, select
Trueto indicate that the control ran as expected. - Click Save.
The QCs will now be visible on the individual Edit Library page.
- On the Libraries list page, enter your project name in the search box and press the
Enterkey on your keyboard. - Click one of the libraries you previously selected.
- Scroll down to the QCs section. The QC you just entered should appear in the table.
4.2 QC status
QC Status describes the quality of a library and may be used to indicate whether it is good enough for sequencing.
- From the Libraries page, find one of your libraries using the search box and click the library name or alias link.
- Change QC passed from
Not ReadytoReady. - Click Save.
4.3 Low Quality Sequencing
Libraries can be marked as having low sequencing quality, which will be shown after the Run exercises.
5. Scanning libraries into your outbox
Finally, update your outbox with your samples/libraries/pools to pass them on to the next team or to put them into storage.
If you haven't already created an outbox, create one now:
- From the navigation panel, choose Boxes.
- Click Add at the top right of the table.
- In the Create Boxes dialog, leave the Quantity at 1 and click Create.
- On the Create Box page, enter the following information:
- Alias:
XX Outbox, replacingXXwith your own initials. Record this name in your worksheet.
- Description: enter a human description of the box purpose
- Box Use: The contents of the box. Select an appropriate use for your entities. Record the
use in your worksheet.

- Box Size: Select
8x12 Storage (scannable).
- Alias:
- Click Save.
Add each of the samples/libraries/pools that you created to the box.
6. Propagating libraries to library aliquots
A library cannot be directly loaded into a lane in a sequencing container (flowcell/SMRTcell) in MISO. A library aliquot must be made and then many aliquots (or just one) can be mixed into a pool for sequencing.
Sequencing Orders are requests for sequencing pools a certain number of times. They are used to keep track of sequencing progress for project management and book-keeping.
6.1 Bulk creating Library Aliquots
Library aliquots can be made in bulk from libraries.
In this exercise, we will create library aliquots from the libraries we made previously.
- On the Libraries page, check all five of the libraries you created.
- From the toolbar, select Make aliquots. Click Create in the dialog.
- Note that the Make aliquots option is also available after bulk creating or bulk editing libraries.
- Enter the library aliquot information:
- QC Status:
Ready - Conc.: Enter any number you wish.
- Conc. Units:
ng/µL - Creation Date: Select the current date.
- Kit: Select any.
-
Kit Lot: Enter today's date, e.g.
2021-06-15 - Targeted Sequencing: Select any if there are options available.
- QC Status:
- Click Save.
7. Creating pool orders
A pool order indicates that a set of library aliquots is ready to be pooled and sequenced. If you hand off library aliquots to another person or team to create the pool, you should create a pool order to pass on the requirements.
7.1 Creating a pool order
- On the Library Aliquots list page, check the five aliquots you just created.
- From the toolbar, select Create Order.
- Enter the order information:
- Alias: Enter the project short name followed by ` Order 1
(e.g.PROJ Order 1`) - Purpose:
Production - Specify sequencing requirements?: Yes
- Platform:
Illumina - Instrument Model:
Illumina HiSeq 2500 - Container Model:
HiSeq PE Flow Cell v4 - Sequencing Parameters:
V4 2×101 - Partitions required: 1
- Draft: No
- Alias: Enter the project short name followed by ` Order 1
- Click Save. Record the order alias in your worksheet.

8. Creating Pools
Pools are the last step for libraries before sequencing and represent the entity that is loaded onto the flowcell lane or SMRTcell. A "pool" can have one or more library aliquots in it. Every lane of sequencing contains only one pool.
8.1 Bulk creating a Pool
Here we will pool all of the library aliquots we added previously to make a single pool.
- On the Library Aliquots page, check the five library aliquots you created.
- From the toolbar at the top left, click Pool Together. Click Create in the dialog.
- Enter the pool information:
- Alias: A short description of the pool contents. Enter the project
short name followed by
_POOL(e.g.PROJ_POOL) - Description: A longer free-text description of the pool.
- Creation Date: Select a date.
- Concentration: Enter a concentration.
- Conc. Units:
ng/µL - Volume: Enter a pool volume.
- Vol. Units:
µL - QC Status:
Ready
- Alias: A short description of the pool contents. Enter the project
short name followed by
- Click Save. Record the Pool alias in your worksheet.

9. Ordering sequencing
Sequencing orders are created on the pool to be sequenced, and include the quantity of sequencing required (counted in lanes/SMRT cells), and the sequencing chemistry required (for Illumina).
9.1 Creating a Sequencing Order
- From the navigation menu, select Pools.
- On the Pools page, select the pool you just created.
- From the toolbar, click Create Orders.
- Enter the order information:
- Purpose: Select any.
- Instrument Model: Select the instrument for sequencing:
Illumina HiSeq 2500. - Container Model: Select
HiSeq PE Flow Cell v4. - Sequencing Parameters: Select
V4 2×101chemistry. - Partitions: the number of lanes that should run for this pool. Enter
2.
- Click Save.
- From the navigation menu, select Pools again.
- Click on the pool you just added a sequencing order to.
- The order will now be visible in the Requested Sequencing Orders section.
- The outstanding sequencing required will be visible under Sequencing Order Status.
The language will vary depending on the platform (i.e., PacBio containers will show SMRTcell Count instead of Lane Count).
9.2 Checking for unfulfilled sequencing orders
The Outstanding Sequencing Orders page is used to decide what needs to be sequenced.
- From the navigation menu, choose Sequencing Orders - Outstanding.
- Verify that the pool you just created is listed.
When enough lanes have been sequenced, the row will disappear from the Outstanding Sequencing Orders page, but remain on the All Sequencing Orders page. Lanes currently being sequenced will be marked as in-progress and remain on the Outstanding Sequencing Orders page until the run transitions to Completed. Columns on the All Sequencing Orders page will disappear if there are no entries (e.g., the Failed column will not be shown if there are no failed runs).
A pool can have many sequencing orders. Sequencing orders for the same platform and chemistry are summed when displayed on the order pages.
Troubleshooting
How do I correct an index on a library?
The index can be changed on either the individual library page or the bulk edit page.
- From the Libraries page, click the name or alias of the library that needs to be changed.
- If necessary, change Index Family. If selected, the indices will be erased.
- Select the correct index under Indices. If the index family supports dual barcoding, another drop down will appear.
What if I forget to put a library aliquot in a pool?
If the library aliquot has not been created:
- From the Libraries page, check the library that needs to be added.
- From the toolbar, click Make aliquots.
- Fill out the new row in the table, then click Save.
Once the library aliquot exists:
- From the Pools page, select the pool that needs the additional library aliquot.
- In the Library Aliquots section, click the
Addbutton. - Enter the library aliquot name, alias, or barcode in the search dialog and click Search.
- Select the correct aliquot in the list and click Next.
- Leave the default proportion of
1and click Add. - Click the Save button at the top right of the page.
How do I make bulk sequencing orders?
Making sequencing orders in bulk can be done from the Pools page:
- Select all the pools needing sequencing.
- Click Create Orders and enter the order information for each pool.
How do I change the targeted sequencing type on a library?
Targeted sequencing is connected to the library aliquot since the same library can be used for multiple targeted sequencing panels.
- From the Library Aliquots page, check the library aliquots that need to be changed.
- From the toolbar, click Edit.
- Select a new Targeted Sequencing from the list.
- Click Save.
How can I add a new targeted sequencing type, kit, or anything else in drop-down menus?
Please email gsi@oicr.on.ca or file a JIRA ticket in GSI Common to get assistance from GSI.
How are matrix tube barcodes assigned to tubes?
Barcodes can be assigned on an individual edit page or in bulk.
To change a single library:
- From Libraries page, click the name or alias of the library that needs to be changed.
- Clear the field labelled Matrix Barcode.
- Scan the new barcode.
- Click Save.
To change many libraries:
- From Libraries page, check the box beside each library that needs to be changed.
- From the toolbar, click Edit.
- Select the cell in the Matrix Barcode column for the library and scan the barcode.
- Repeat for all the libraries.
- Click Save.
How can I delete a library?
Libraries can be deleted from the Libraries list page by selecting the libraries and clicking Delete. A library cannot be deleted if it has any aliquots. Only the library's creator or an admin can delete a library. Deleted items are listed on the Deletions list page. Deleting a library is permanent and cannot be undone.
How can I delete a library aliquot?
Library aliquots can be deleted from the Library Aliquots list page by selecting the aliquots and clicking Delete. A library aliquot cannot be deleted if it has been added to any pools. Only the aliquot's creator or an admin can delete an aliquot. Deleted items are listed on the Deletions list page. Deleting a library aliquot is permanent and cannot be undone.
How can I delete a pool?
Pools can be deleted from the Pools list page by selecting the pools and clicking Delete. A pool cannot be deleted if it has been added to any partitions. Any associated sequencing orders will be deleted along with a pool. Only the pool's creator or an admin can delete a pool. Deleted items are listed on the Deletions list page. Deleting a pool is permanent and cannot be undone.
How can I delete a sequencing order?
Sequencing orders can be deleted from the Edit Pool page by selecting the orders and clicking Delete. Deleted items are listed on the Deletions list page. Deleting a sequencing order is permanent and cannot be undone.
What if I assign a library to the wrong parent aliquot?
You can delete the library and re-create it. If this is not appropriate because the library is already included in a sequencer run, please email gsi@oicr.on.ca or file a JIRA ticket in GSI Common to get assistance from GSI.
| < Samples | Home | Sequencing > |
 MISO
MISO